On June 22-26, 2020, as part of the project „Oxford Nanopore Technology: Enzyme Optimization and Genomic Data Analysis for Commercial Applications”, a DNA sequencing experiment using Oxford Nanopore technology was conducted at the Rzeszów University of Technology. The experiment was divided into two parts - experimental and bioinformatic, implemented respectively in the Department of Biotechnology and Bioinformatics of the Faculty of Chemistry and in the Department of Complex Systems of the Faculty of Electrical and Computer Engineering. The efficient sequencing as well as correct analysis of the obtained data was supervised by Jan Gawor from the Laboratory of DNA Sequencing and Oligonucleotide Synthesis at the Institute of Biochemistry and Biophysics of the Polish Academy of Sciences. The first part of the experiment involved mainly: PhD Marta Sochacka-Piętal, PhD Michał Pietal, MSc Małgorzata Semik, MSc Michał Wroński. The necessary organizational and technical assistance was also provided by Prof. Mirosław Tyrka. In the second part, bioinformatic, it was required to involve the rest of the project team, including PhD Dominik Strzałka, MSc Magdalena Totoń, Eng. Michał Ćmil and MSc Anna Czmil. The experiment lasted 5 working days: the first two days concerned work in a wet laboratory, the remaining days were intended for bioinformatic works, including data analysis and their processing on the servers of the Department of Complex Systems.
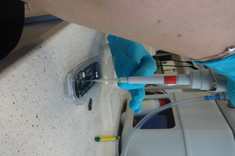
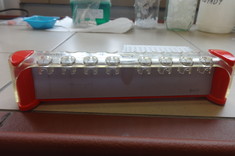
The starting material in the experimental part was DNA from thermotolerant and thermophilic strains of Bacillus genus from the collection of the Department of Biotechnology and Bioinformatics PRz. During the experimental work, project participants became acquainted with procedures for appropriate DNA isolation and preparation of genomic libraries, which are then applied to the latest generation sequencer - MinION. The sequencing was a complete success and the obtained raw data was further processed by IT.
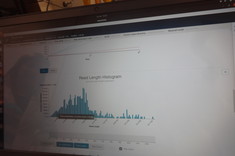
During the bioinformatic part of the experiment, the project contractors got acquainted with a number of bioinformatic tools (including Bandage, Guppy, NanoPlot, Fly, Canu) used in de novo assembly of bacterial genomes. Appropriate computing power and remote access to server resources were required. In addition, hybrid genomic assembly was performed using genomic data from Oxford Nanopore and Illumina sequencing. As a result of the experiment, de novo complete genomic sequences of the analyzed Bacillus sp. Strains were obtained.
Further similar experiments involving sequencing of mutations in cultured bacteria will be carried out later in the project. Depending on the successively obtained results, the conditions of the series of experiments carried out will be changed accordingly. This should finally lead to the preparation of the bioinformatics server implementing pipelined processing of the obtained data and enable optimization of this process.